Research
Research in the group of Prof. Petra Imhof
Our research aims at understanding chemical and physical processes in biological macromolecules such as proteins and enzymes by means of computer simulations.
At finite, e.g. physiological temperature, molecules are not static entities with a fixed geometry but rather exhibit dynamics, occupying and interchanging between several conformational and configurational states. Often, only a few of those states render the molecule functional, such as the folded form of a protein. And even so often, the transition between states is associated with function such as the switching in a signalling protein, or the chemical reaction catalysed by an enzyme. Both, the intrinsic probability to populate different states as well as the probabilities for transitions between them are influenced by the physical conditions and the molecules’ interactions with their environment. Molecular simulations allow us explore the underlying interplay of dynamics, interaction, and reactivity.
We are targeting the rational development of biomimetic substitutes for enzymes (catalysts), the design of specific inhibitors and rational protein engineering, using molecular dynamics simulations and combined QM/MM approaches. Current applications are protein-substrate interaction and substrate recognition, exploring enzymatic reaction pathways and enzymatic catalysis as well as photoinduced transformations from conformational transitions to reactions.
Protein-DNA Interactions
DNA repair
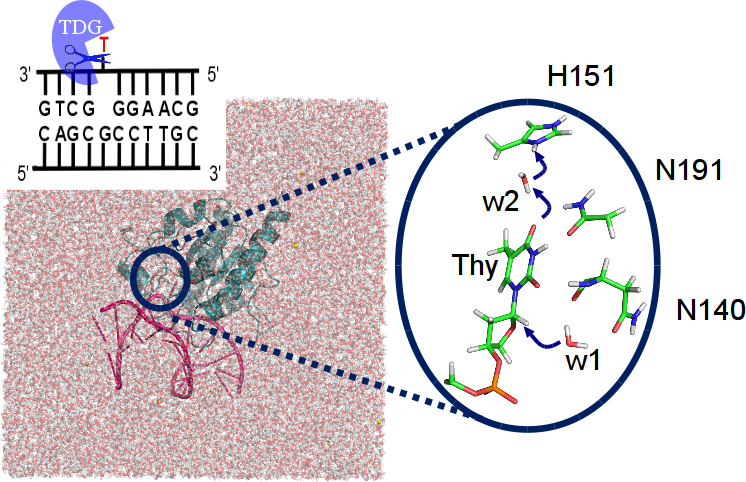
Our DNA is constantly threatened by damages due to e.g. UV radiation or chemical agents. We study the recognition and removal of the damaged or mismatched site by molecular dynamics simulations and QM/MM calculations [30][34][35][44]
Sequence recognition
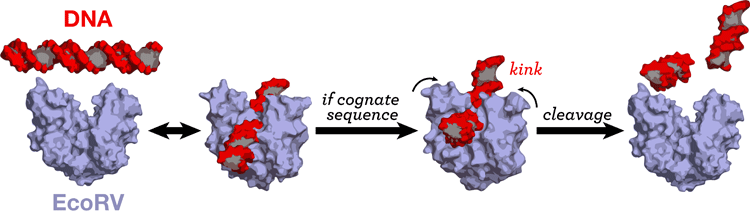
Most DNA-processing proteins act at a specific sequence or target site of the DNA. We are investigating the protein-DNA interaction in cognate and non-cognate complexes so as to understand specificity. [23] [26] [38] [48].
Compex transitions and reaction mechanisms
Transition Networks
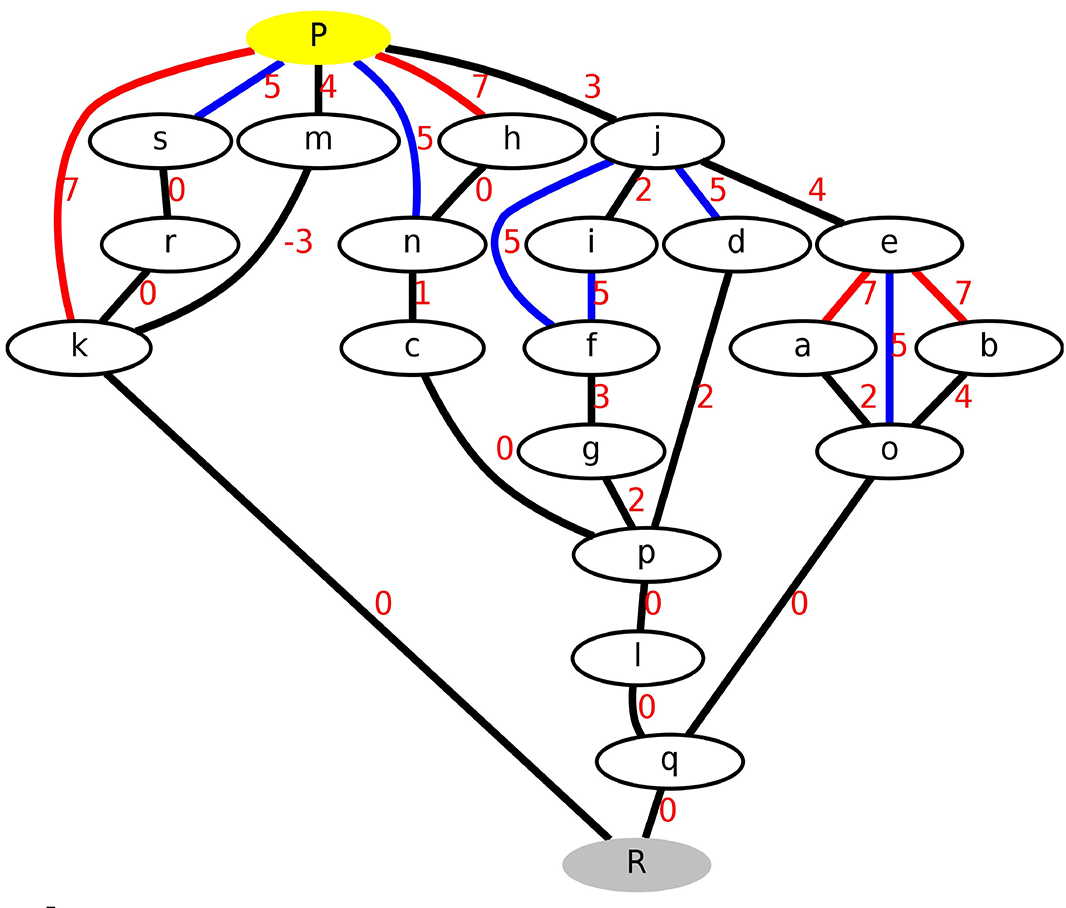
Complex transitions in macromolecules such as enzymatic reactions usually cannot be described by a simple order parameter, a single reaction coordinate, or a single minimum energy pathway. Transition networks are a discretised representation of the energy surface, that describes all the relevant conformations and chemical states of the system as well as the transition pathways and barriers connecting them, allowing to sample possible transition (reaction) pathways in high-dimensional systems without an a priori definition of a specific reaction coordinate [36] [37] [45]. Appliocations are e.g. proton transfer in membrane proteins or enzymatic mechanisms.
Enzymatic mechanisms
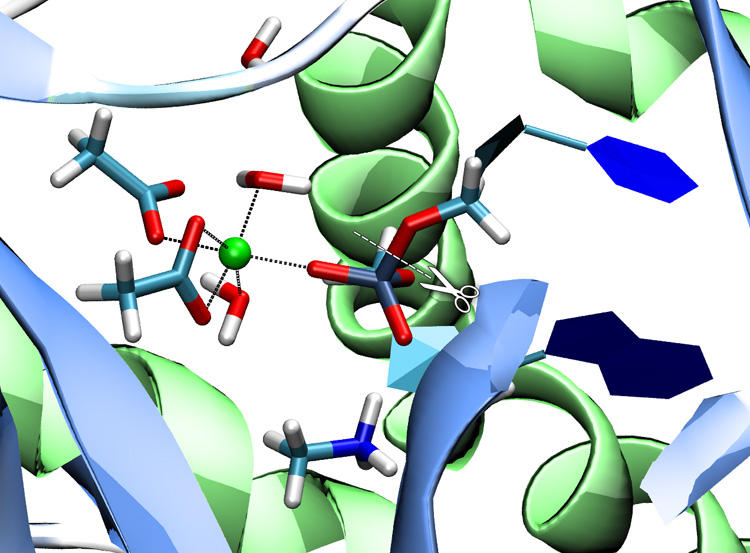
Phosphate transfer reactions are essential to many biochemical processes including nucleotide synthesis, signal transduction, and cell growth regulation. Despite their stability against hydrolysis, phosphate esters can be processed by phosphate transferring enzymes under mild (physiological) conditions in minutes or even seconds. We are investigating the reaction mechanisms of a variety of phosphate transferring enzymes (nucleases, kinases, phosphatases) applying QM/MM methods. [22] [41] [46]
Communication and signalling
Often molecular function is controlled through large distances within the same molecule or in a molecular complex. To analyse allosteric pathways and signalling we construct communication networks. The system of interest, e.g. protein, is represented by a weighted graph, where individual residues correspond to the network nodes. Edges connecting pairs of nodes are weighted corresponding to different interaction metrics representing communication. Graph-theoretical analysis is then applied to determine e.g. shortest communication pathways, community structures in the network, and the importance of particular residues or domains in communication [40][42].
Computational Spectroscopy
Vibrational dynamics
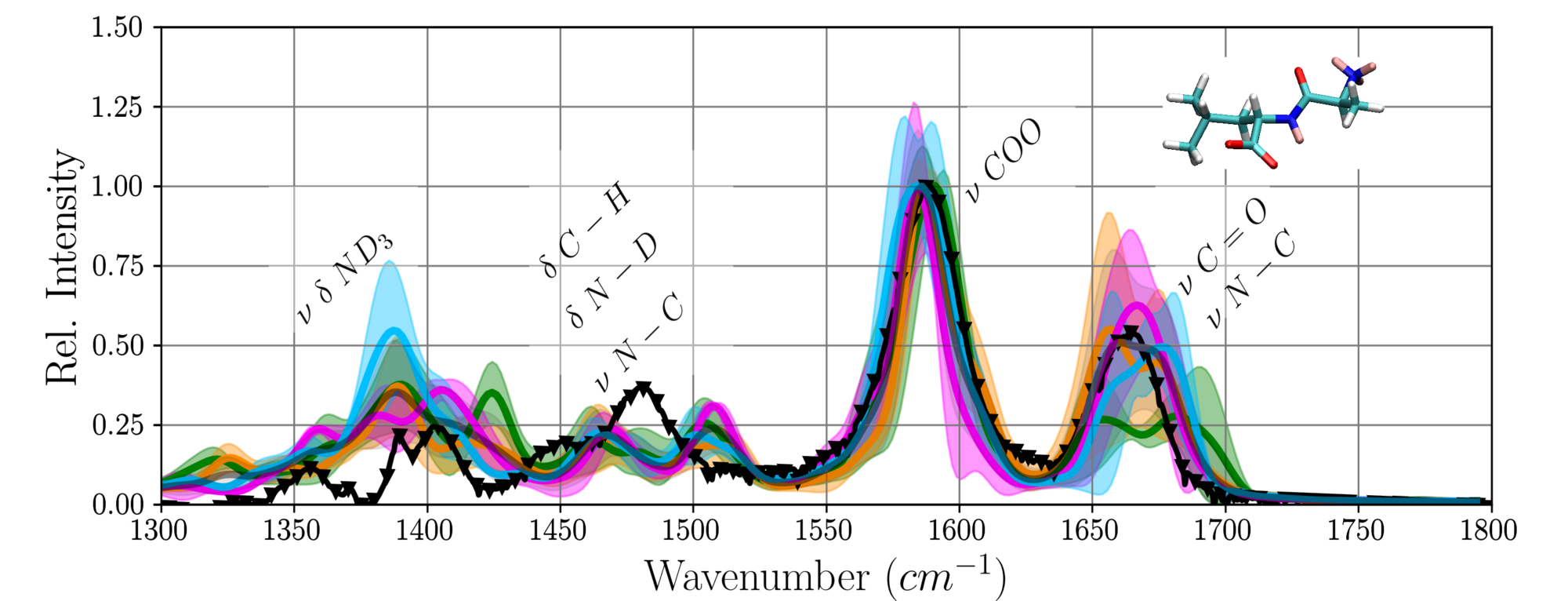
From a classical molecular dynamics simulation the most probable conformations and the dominant dynamics of the small peptide Ala-Leu could be elucidated. Infrared-spectra computed from first-principles MD calculations of representative conformations agree well with the experimental spectrum [43].
Fluorescent Proteins
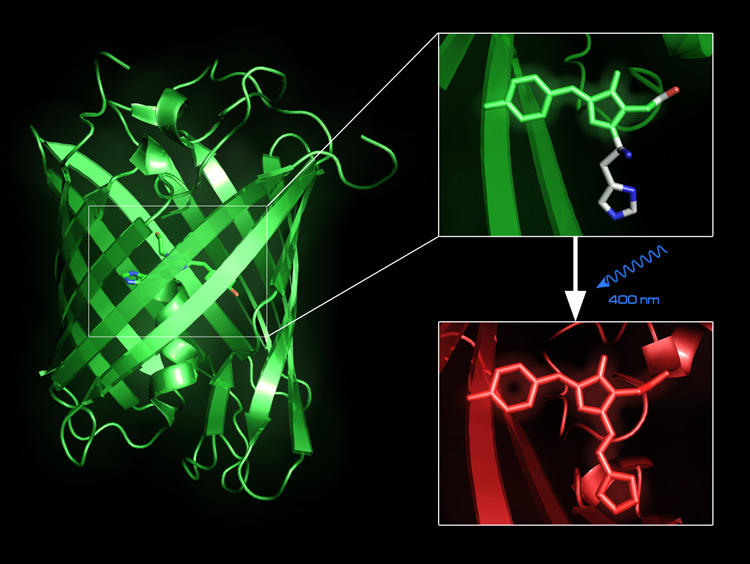
Fluorescent proteins, such as the Green Fluorescent Protein, can be used to trace intracellular processes in vivo. Some of these, e.g. EosFP, are photo-convertible. Our QM/MM calculations confirmed the proposal of the protein’s neutral chromophore state to be the starting point for photo-induced red-to-green conversion [29]. We are conducting simulation studies in order to better understand the relationship between structure and fluorescent properties.
